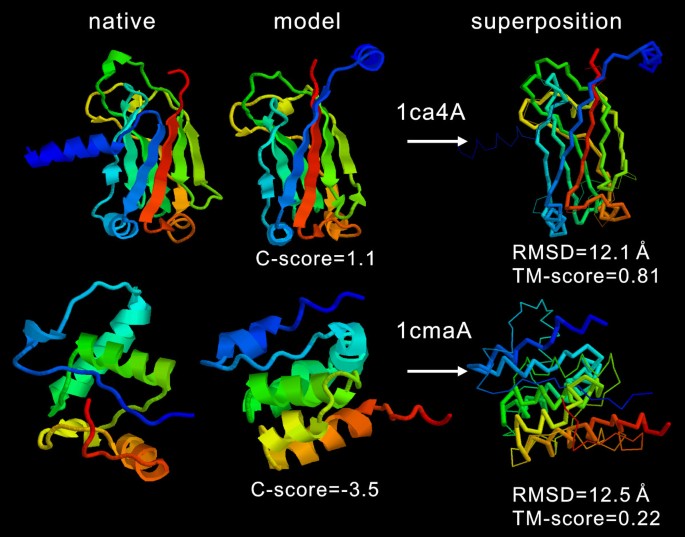
I‐TASSER: Fully automated protein structure prediction in CASP8 - Zhang - 2009 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
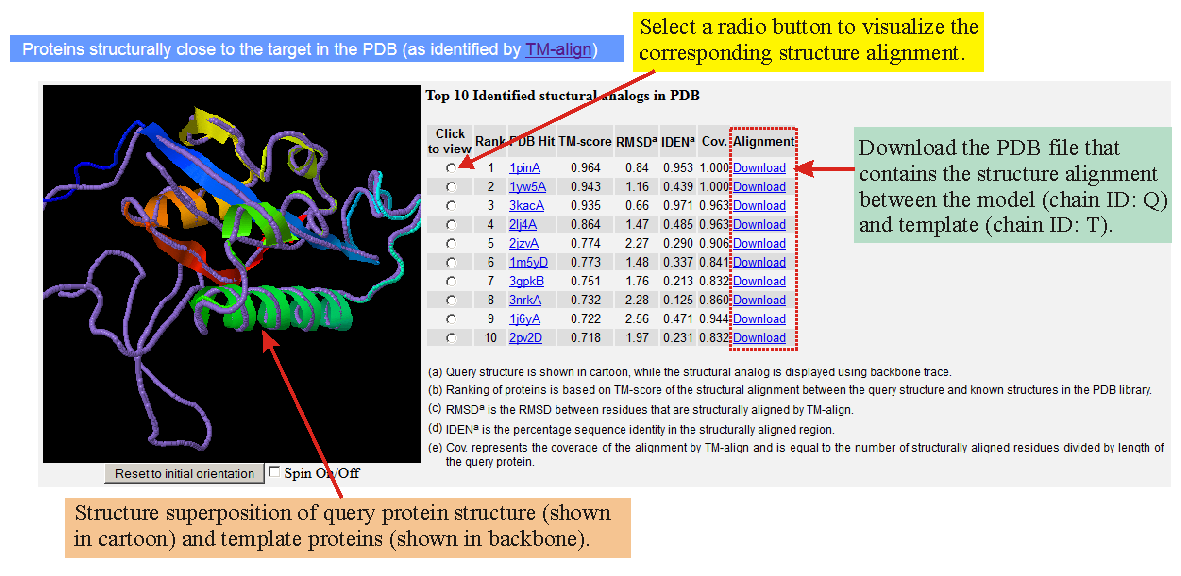
![PDF] I-TASSER server: new development for protein structure and function predictions | Semantic Scholar PDF] I-TASSER server: new development for protein structure and function predictions | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/682fc6a2dcf6edaadacb1a2f90fdd137aef28d15/5-Figure4-1.png)
PDF] I-TASSER server: new development for protein structure and function predictions | Semantic Scholar

Exploring the antihyperglycemic potential of tetrapeptides devised from AdMc1 via different receptor proteins inhibition using in silico approaches - Ghulam Mustafa, Hafiza S Mahrosh, Muddassar Zafar, Syed A Attique, Rawaba Arif, 2022
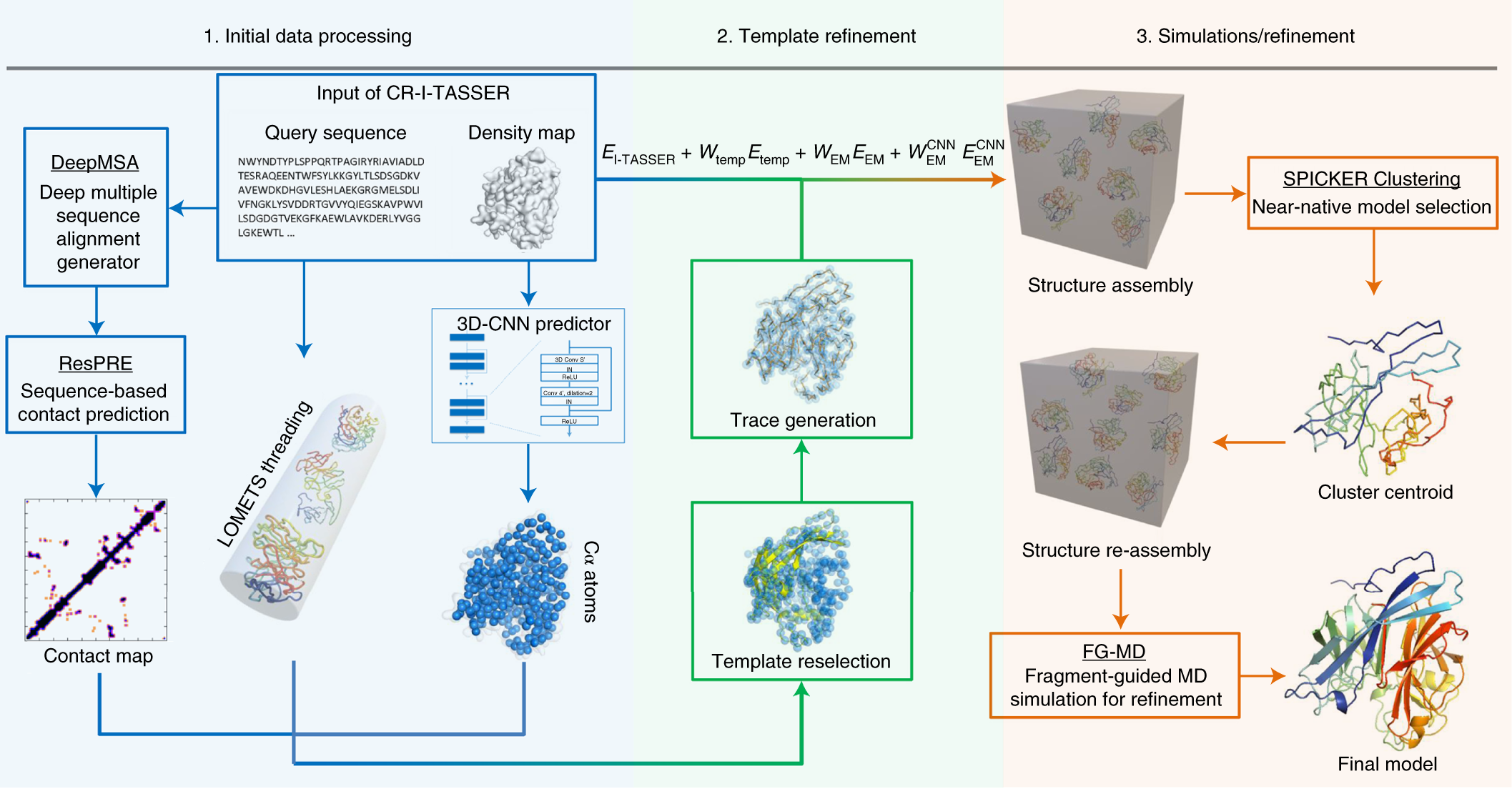
CR-I-TASSER: assemble protein structures from cryo-EM density maps using deep convolutional neural networks | Nature Methods

Template‐based modeling and free modeling by I‐TASSER in CASP7 - Zhang - 2007 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
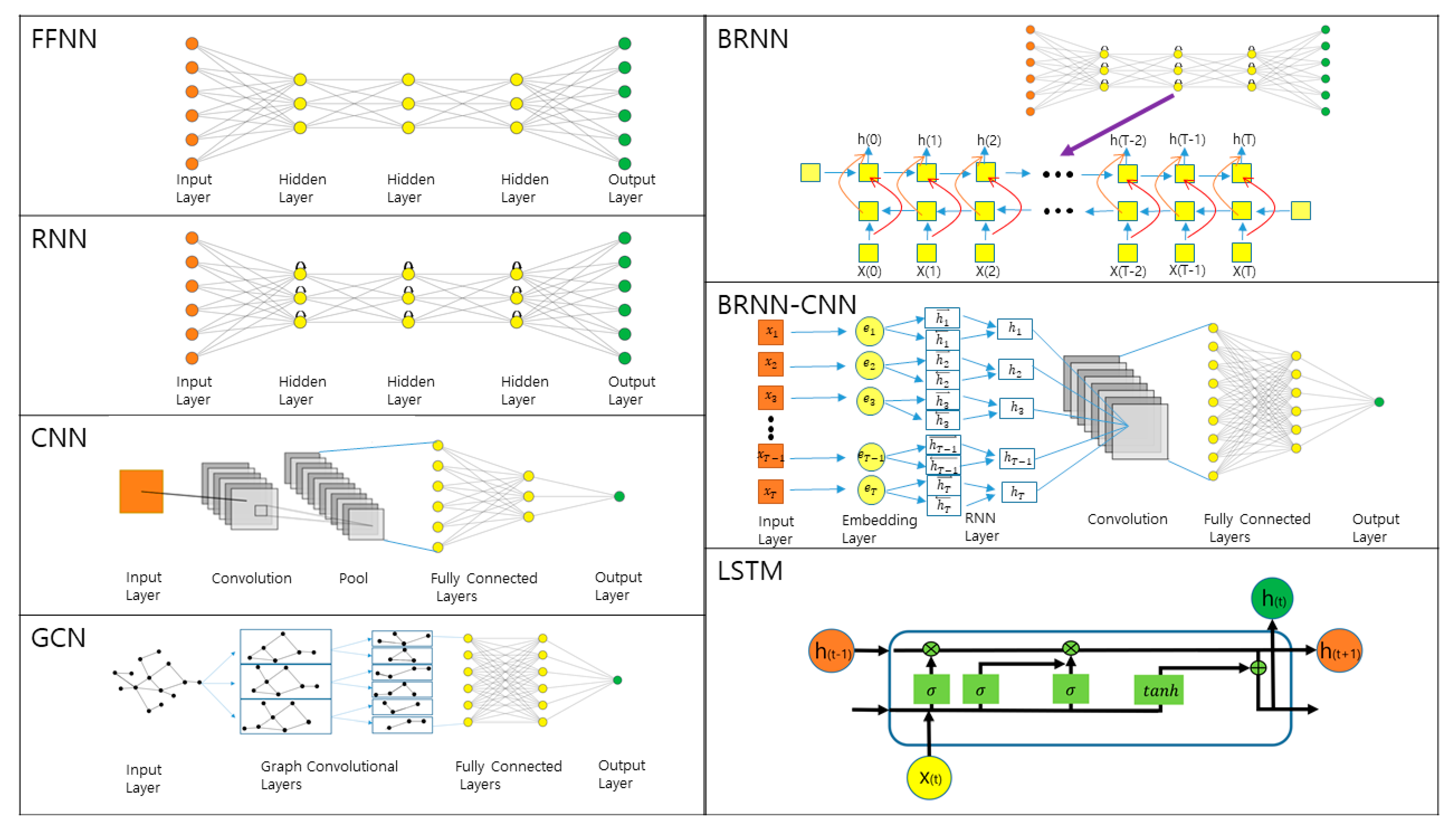
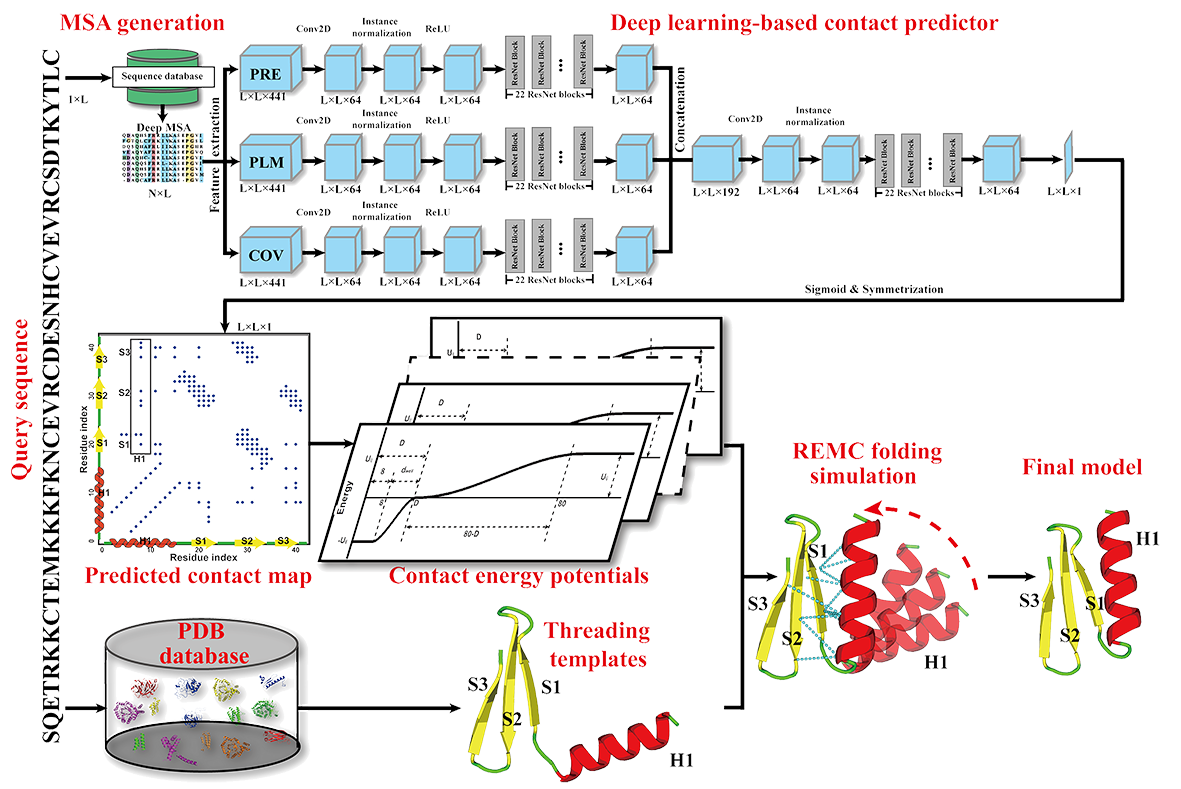
IJMS | Free Full-Text | Recent Applications of Deep Learning Methods on Evolution- and Contact-Based Protein Structure Prediction

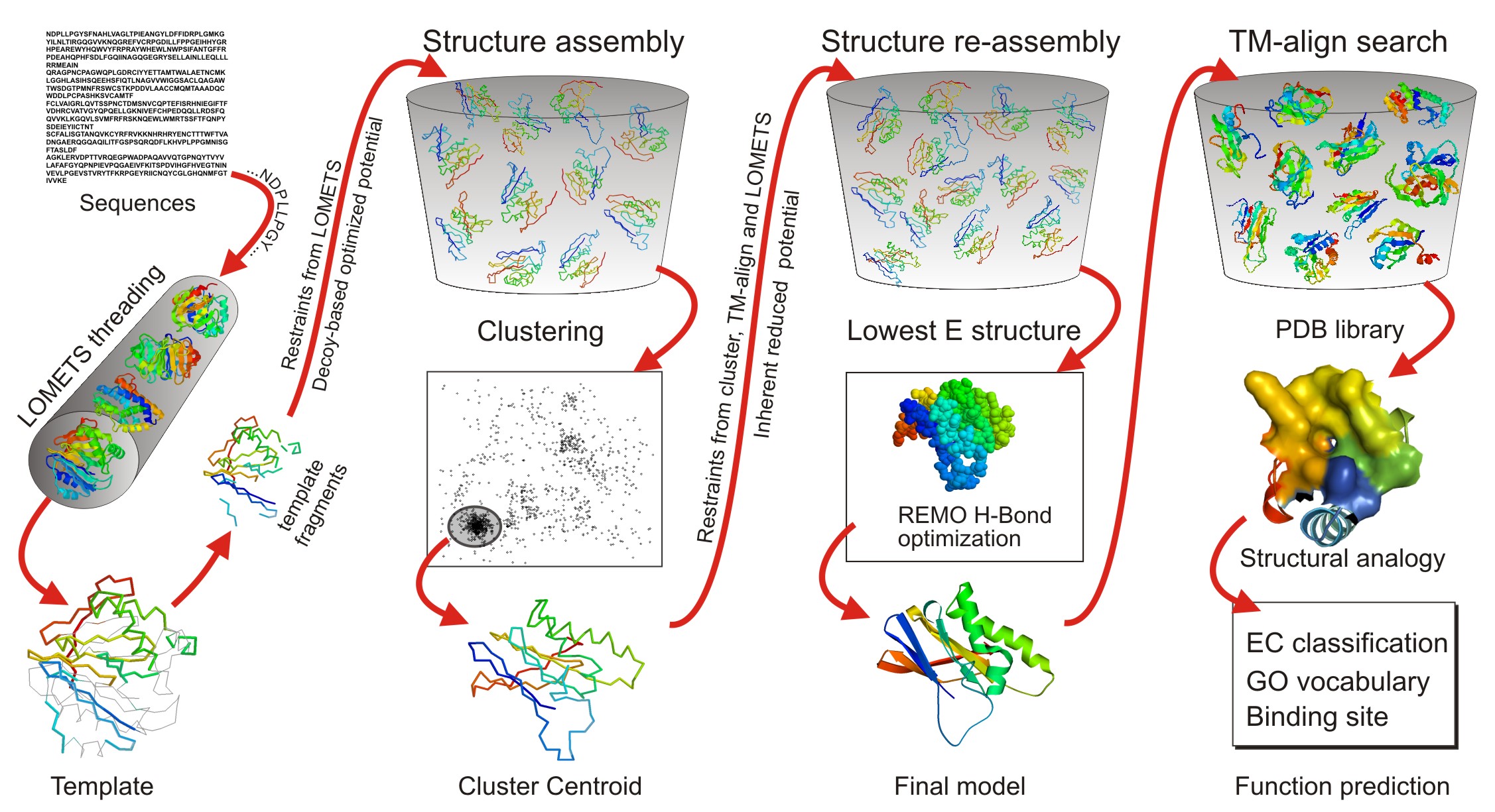
I-TASSER-MTD: a deep-learning-based platform for multi-domain protein structure and function prediction | Nature Protocols

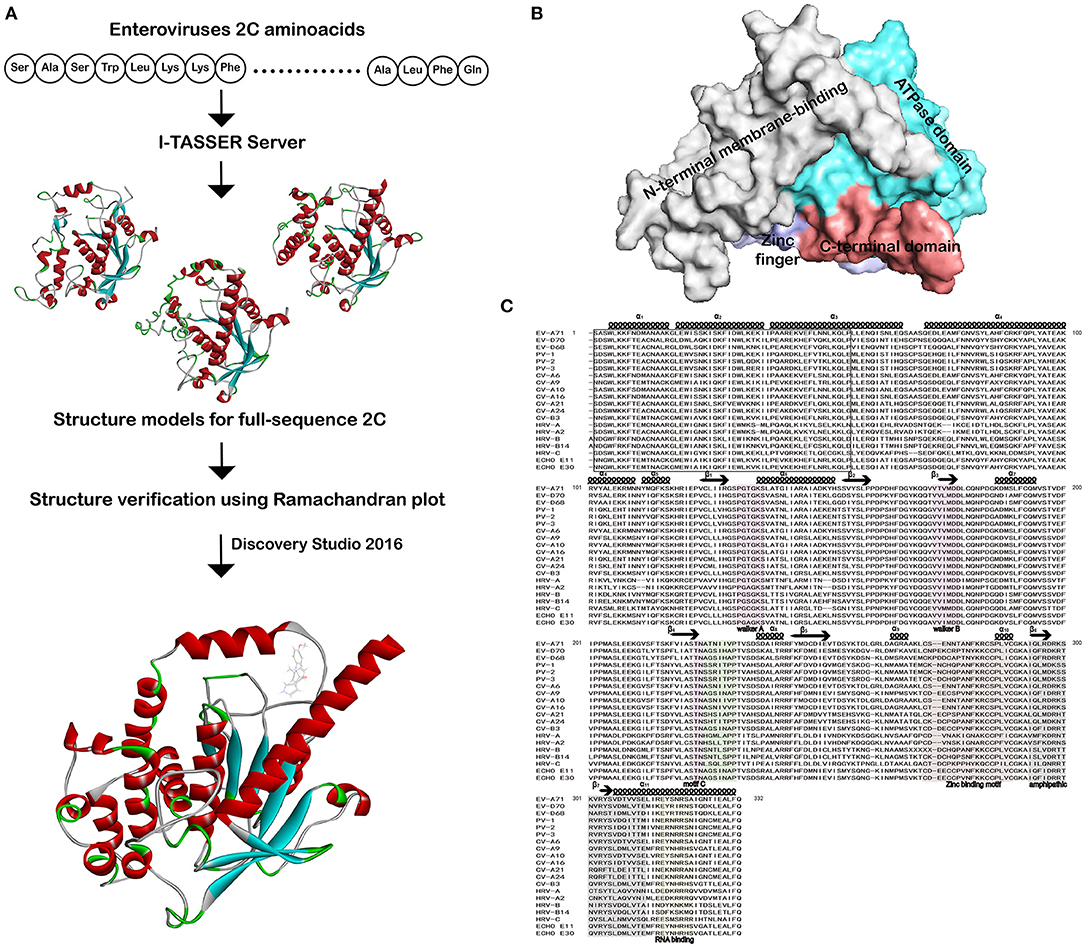
A) Tertiary structure (3D) of the Protein Model-54, generated by: a)... | Download Scientific Diagram

Automated protein structure modeling in CASP9 by I‐TASSER pipeline combined with QUARK‐based ab initio folding and FG‐MD‐based structure refinement - Xu - 2011 - Proteins: Structure, Function, and Bioinformatics - Wiley Online


![General workflow of I-TASSER for protein structure prediction [30] | Download Scientific Diagram General workflow of I-TASSER for protein structure prediction [30] | Download Scientific Diagram](https://www.researchgate.net/publication/281540998/figure/fig1/AS:281366399864834@1444094385414/General-workflow-of-I-TASSER-for-protein-structure-prediction-30.png)